Evaluating and Improving Heritability Models using Summary Statistics
BiRC member Doug Speed, in collaboration with David Balding and John Holmes (University of Melbourne), has recently developed a new tool that improves our understanding of the genetic factors underlying complex traits. Details of the tool have been published this week in the journal Nature Genetics.
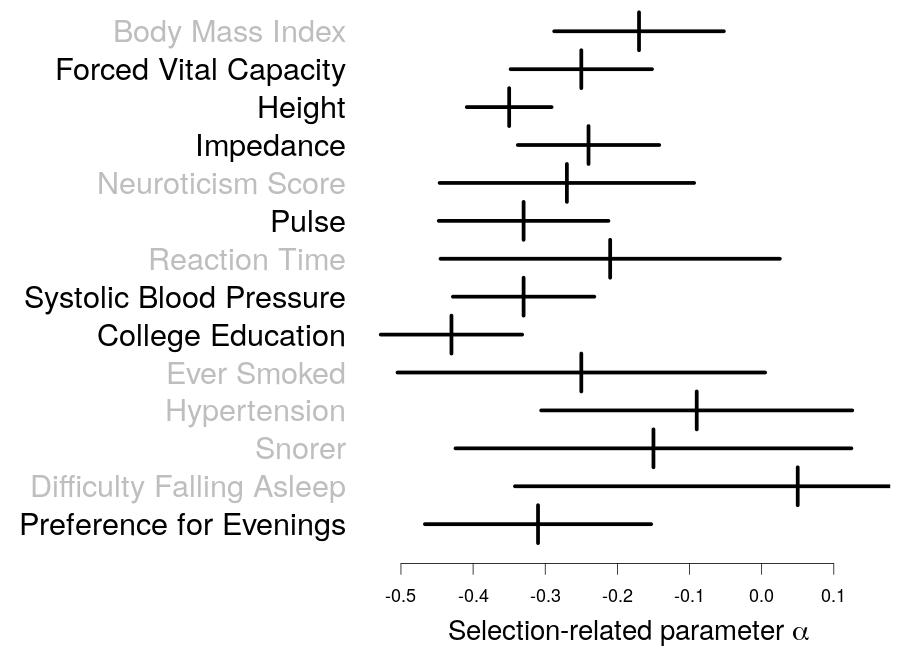
The human genome contains millions of genetic variants, the most simple of which are single nucleotide polymorphisms (SNPs). These SNPs influence a wide range of complex traits. For example, they determine how likely we are to develop psychiatric traits such as schizophrenia, autoimmune traits such as Crohn's Disease, or cardiovascular traits such as heart disease. When we analyze SNP data, it is often necessary to specify a "heritability model", which describes in advance how much we think each SNP will contribute. It turns out that for many analyses, the choice of heritability model is very important (i.e., if we change the heritability model, it substantially changes the results of the analysis). In recent years, many different heritability models have been proposed, but until now, there has been no way to decide which of these is the best one to use.
In his current work, Doug presents a tool for evaluating and improving heritability models. He first uses this tool to determine which existing heritability model is best, then to create a new model that is more realistic than any previously proposed. Finally, he uses this new heritability model to improve our understanding of 31 complex traits; for example, to quantify how much different types of SNPs influence different traits, and to measure the impact of natural selection.
According to Doug, "The new tool not only improves our understanding of complex traits now, but will also enable us to more efficiently analyze future data." With this in mind, Doug has integrated the new tool into his software package LDAK (www.ldak.org), so that it is freely available for other researchers to apply to their own data. For more details, see the article "Evaluating and Improving Heritability Models using Summary Statistics".
Read the article 'Evaluating and improving heritability models using summary statistics' here.
